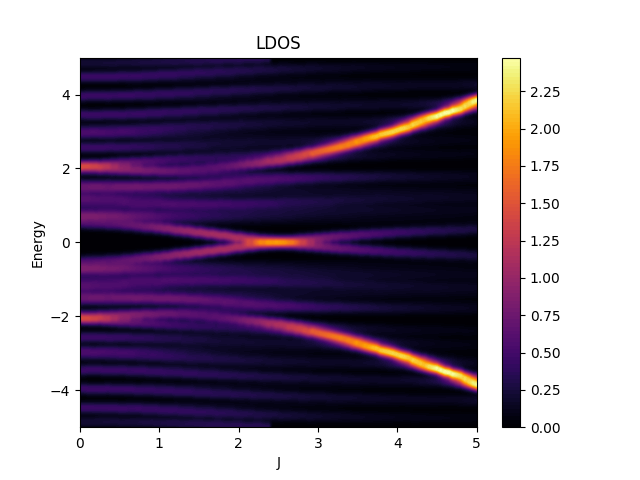
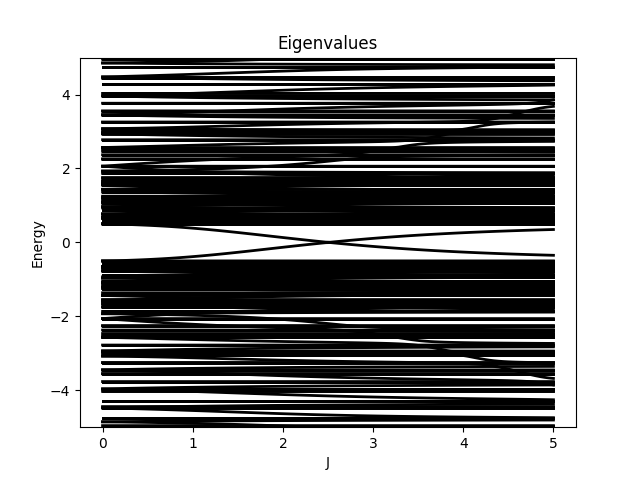
where \(\mathbf{I}\) is the impurity site.
#include "TBTK/PropertyExtractor/Diagonalizer.h"
#include <complex>
using namespace std;
using namespace TBTK;
using namespace Visualization::MatPlotLib;
complex<double> i(0, 1);
public:
complex<double> getHoppingAmplitude(
) const{
return J*(1. - 2*spin)*(1. - 2*particleHole);
}
void setJ(complex<double> J){
this->J = J;
}
private:
complex<double> J;
};
int main(){
const unsigned int SIZE_X = 11;
const unsigned int SIZE_Y = 11;
const double t = -1;
const double mu = -2;
const double Delta = 0.5;
JCallback jCallback;
for(unsigned int x = 0; x < SIZE_X; x++){
for(unsigned int y = 0; y < SIZE_Y; y++){
for(unsigned int spin = 0; spin < 2; spin++){
for(unsigned int ph = 0; ph < 2; ph++){
-mu*(1. - 2*ph),
{x, y, spin, ph},
{x, y, spin, ph}
);
if(x+1 < SIZE_X){
t*(1. - 2*ph),
{x+1, y, spin, ph},
{x, y, spin, ph}
) + HC;
}
if(y+1 < SIZE_Y){
t*(1. - 2*ph),
{x, y+1, spin, ph},
{x, y, spin, ph}
) + HC;
}
}
Delta*(1. - 2*spin),
{x, y, spin, 0},
{x, y, (spin+1)%2, 1}
) + HC;
}
}
}
for(unsigned int spin = 0; spin < 2; spin++){
for(unsigned int ph = 0; ph < 2; ph++){
jCallback,
{SIZE_X/2, SIZE_Y/2, spin, ph},
{SIZE_X/2, SIZE_Y/2, spin, ph}
);
}
}
const unsigned int NUM_ITERATIONS = 100;
NUM_ITERATIONS,
});
Range j(0, 5, NUM_ITERATIONS);
for(unsigned int n = 0; n < NUM_ITERATIONS; n++){
jCallback.setJ(j[n]);
= propertyExtractor.getEigenValues();
const double LOWER_BOUND = -5;
const double UPPER_BOUND = 5;
const unsigned int RESOLUTION = 500;
propertyExtractor.setEnergyWindow(
LOWER_BOUND,
UPPER_BOUND,
RESOLUTION
);
{SIZE_X/2, SIZE_Y/2, IDX_SUM_ALL, IDX_SUM_ALL},
{SIZE_X/4, SIZE_Y/4, IDX_SUM_ALL, IDX_SUM_ALL}
});
const double SMOOTHING_SIGMA = 0.1;
const unsigned int SMOOTHING_WINDOW = 51;
ldos = Smooth::gaussian(
ldos,
SMOOTHING_SIGMA,
SMOOTHING_WINDOW
);
totalLdos[{n, e}] = ldos(
{
SIZE_X/2,
SIZE_Y/2,
IDX_SUM_ALL,
IDX_SUM_ALL
},
e
);
}
for(
unsigned int e = 0; e < eigenValues.
getSize(); e++)
totalEigenValues[{n, e}] = eigenValues(e);
}
Plotter plotter;
plotter.setNumContours(100);
plotter.setAxes({
{0, {0, 5}},
{1, {-5, 5}},
});
plotter.setTitle("LDOS");
plotter.setLabelX("J");
plotter.setLabelY("Energy");
plotter.setBoundsY(-5, 5);
plotter.plot(totalLdos);
plotter.save("figures/LDOS.png");
plotter.clear();
plotter.setTitle("Eigenvalues");
plotter.setLabelX("J");
plotter.setLabelY("Energy");
plotter.setAxes({
{0, {0, 5}},
{1, {-5, 5}}
});
plotter.setBoundsY(-5, 5);
for(
unsigned int e = 0; e < (
unsigned int)model.
getBasisSize(); e++){
plotter.plot(
totalEigenValues.getSlice({_a_, e}),
{{"color", "black"}, {"linestyle", "-"}}
);
}
plotter.save("figures/EigenValues.png");
}